title: Acylation and Deacylation mechanism of Class A β-lactamase mutant E166Y with Cephaloridine
雷金平(Jinping Lei) (TALK)
institution: 香港科技大学(HKUST)
abstract:
β-lactamases confer resistance to β-lactam-based antibiotics. There is great interest in understanding their mechanisms to enable the development of β-lactamase–specific inhibitors. The mechanism of Class A β-lactamases has been studied extensively, revealing Lys73 and Glu166 as general bases that assist the catalytic residue Ser70. However, the specific roles of these two residues within the catalytic cycle remains not fully understood. To help resolve this question, we identified a Glu166Tyr (E166Y) mutant that is functional but kinetically slowed the deacylation. We also carried out time-resolved crystallographic study of a full cycle of the catalytic reaction, and obtained structures that represent apo, ES*-acylation and ES*-deacylation states with antibiotics cephaloridine.
In our simulation work, we aim to characterize the complete mechanism for the acylation and deacylation mechanism of this E166Y β-lactamase mutant by computational simulations, and provide detailed insights into the reaction dynamic. Herein, we use Born-Oppenheimer ab initio Quantum Mechanics/Molecular Mechanics–Molecular Dynamics (QM/MM–MD) simulations with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions. We have also determined the entire free energy profile of the acylation and deacylation reaction.
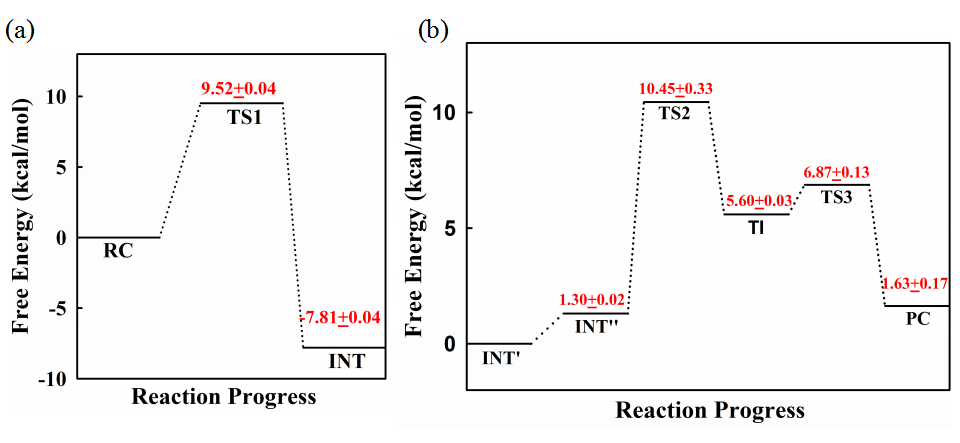
Our calculated results show that the acylation reaction proceeds in a single step, in which the nucleophilic attack of Ser70 and cleavage of the C-N bond of cephaloridine happen in concerted, and the deprotonated Lys73 rather than Tyr166 that act as general base to deprotonate Ser70 (Fig. 1a). For the proton transfer in the acylation reaction, Ser130 acts as proton donor to protonate the nitrogen of the cleaved C-N bond, and the proton transfer from Lys73 to Ser130 can happen spontaneously associated with the acylation of E166Y by cephaloridine. The free energy barrier for the acylation is about 13 kcal/mol (Fig. 2a), agree with experiment results that acylation rate of E166Y is as fast as wide type.
On the other hand, our calculated results reveal that the deacylation reaction proceed in two steps: the nucleophilic attack of deacylation water to form a tetrahedral intermediate state, and the leaving of Ser70 from the ligand CED. In these deacylation steps, the deprotonated Tyr166 acts as general base to deprotonate the deacylation water, and Lys73 donors its proton for the leaving Ser70 (Fig. 1b). Our calculated deacylation mechanism and free energy barrier (Fig. 2b) are similar to class C β-lactamases.1,2
In summary, our computational simulations can provide insight into the development of β-lactamase–specific antibiotic inhibitors.
References
1. Ravi Tripathi and Nisanth N. Nair. J. AM. CHEM. SOC. 2013, 135, 14679−14690.
2. Ravi Tripathi and Nisanth N. Nair. J. PHYS. CHEM. B. 2016, 120, 2681-2690.
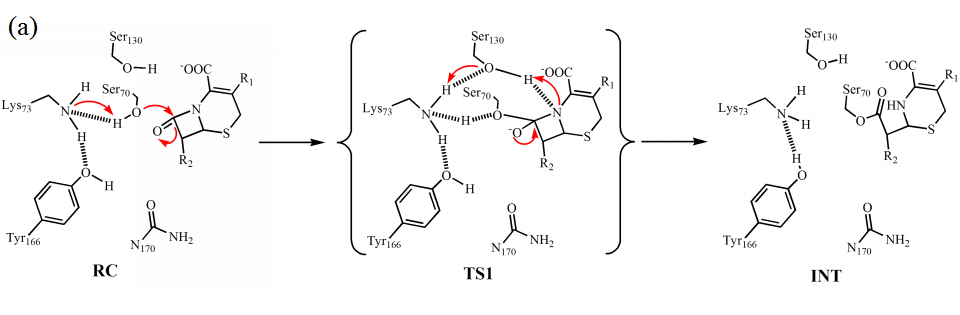

Figure 1. The proposed mechanisms of acylation (a) and deacylation (b) reactions between Class A β-lactamase and cephaloridine as obtained from aiQMMM-MD simulations. The arrows denote the transfer of negative charge. RC, reactant complex; TS1, first transition state; INT, intermediate; TI, tetrahedral intermediate; PC, product complex.