

Multi-Omics Analysis
(1) Next Generation Sequencing techniques generate vast amount of genetic data that requires highly scalable and efficient tools for analyzing these data. Sequencing reads from the machines can be in the range of TB-PB range, sequential algorithms can no longer meet the needs. In our lab, we focus on developing De Bruijn graph based approach for assembling the raw reads. Through mathematical abstraction and building an asynchronous computing model, we have developed SWAP-Assembler for large scale genome assembly. We have demonstrated that SWAp-Assembler can scale to tens of thousands of cores on Mira@Argonne National Lab, and Tianhe-1A@National Supercomputing Center in Tianjin. This is an open source project, and the software can be download from http://sourceforge.net/projects/swapassembler/.
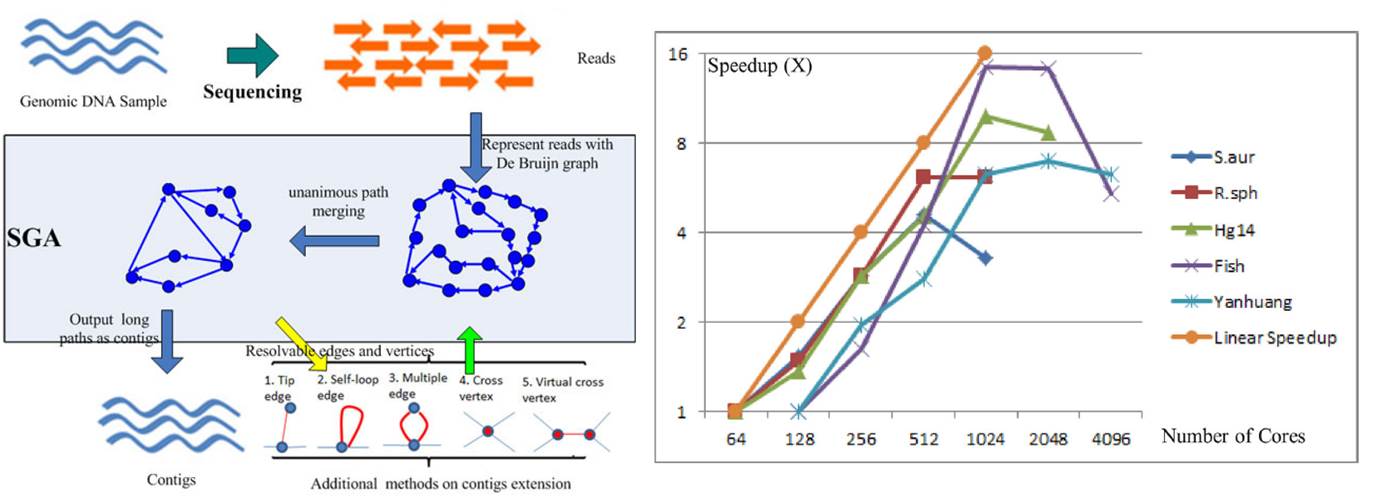 De Bruijn Graph based Genome Assembly and its High Scalability on Tianhe 1A
De Bruijn Graph based Genome Assembly and its High Scalability on Tianhe 1A
(2) CircRNA: Many circRNA transcriptome data were deposited in public resources, but these data show great heterogeneity. Researchers without bioinformatics skills have difficulty in investigating these invaluable data or their own data. Here, we specifically designed circMine (http://hpcc.siat.ac.cn/circmine) that provides 1 821 448 entries formed by 136 871 circRNAs, 87 diseases and 120 circRNA transcriptome datasets of 1107 samples across 31 human body sites. circMine further provides 13 online analytical functions to comprehensively investigate these datasets to evaluate the clinical and biological significance of circRNA. To improve the data applicability, each dataset was standardized and annotated with relevant clinical information. circMine provides userfriendly web interfaces to browse, search, analyze and download data freely, and submit new data for further integration, and it can be an important resource to discover significant circRNA in different diseases. (website:http://hpcc.siat.ac.cn/circmine/home)
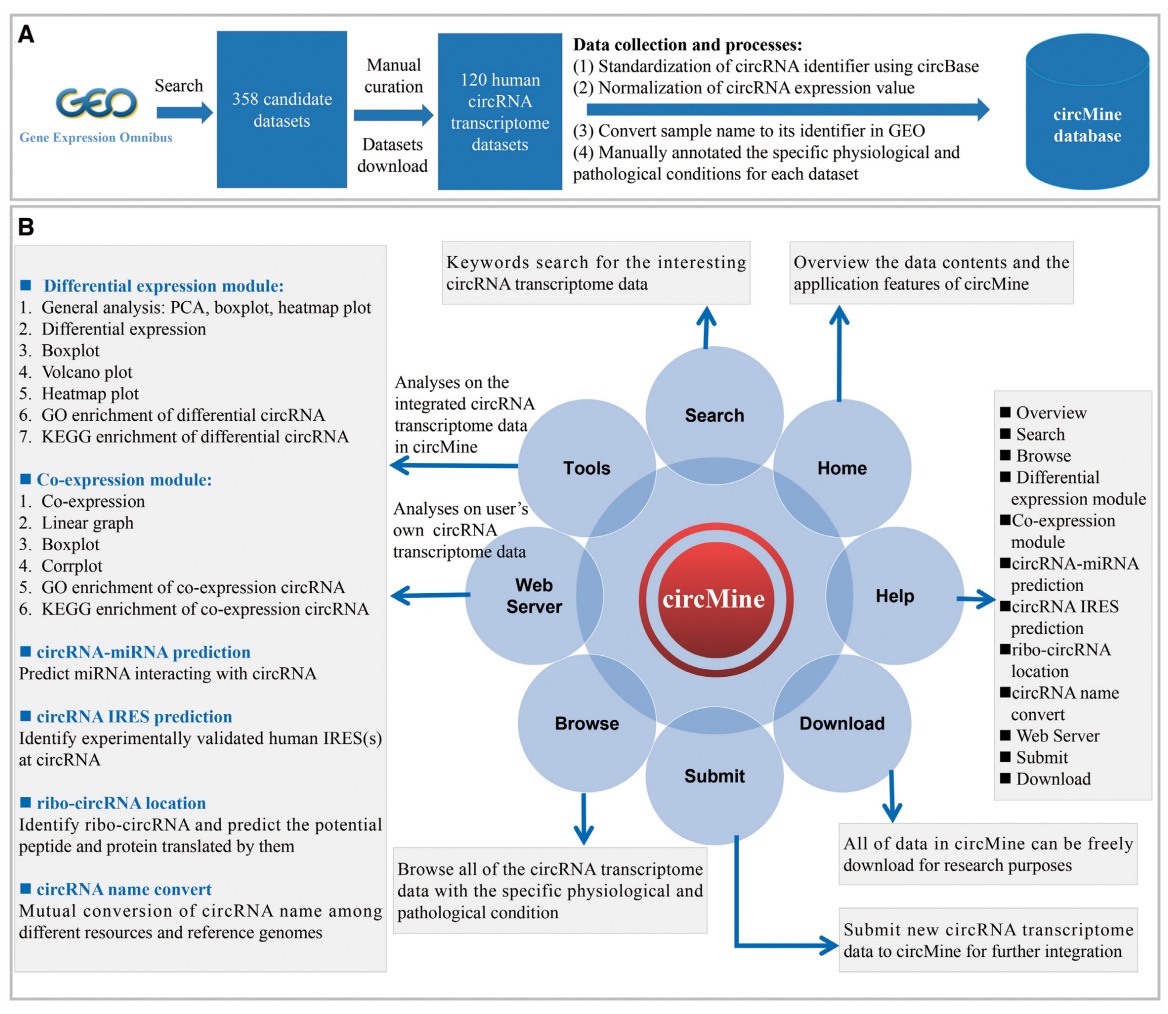 The scheme for data collection and manual curation of circMine
The scheme for data collection and manual curation of circMine
(3)Transcriptomics for drug discovery: Many open access transcriptomic data of coronavirus disease 2019 (COVID-19) were generated, they have great heterogeneity and are difficult to analyze. To utilize these invaluable data for better understanding of COVID-19, additional software should be developed. Especially for researchers without bioinformatic skills, a user-friendly platform is mandatory. We developed the COVID19db platform that provides 39 930 drug-target-pathway interactions and 95 COVID-19 related datasets, which include transcriptomes of 4127 human samples across 13 body sites associated with the exposure of 33 microbes and 33 drugs/agents. To facilitate data application, each dataset was standardized and annotated with rich clinical information. The platform further provides 14 different analytical applications to analyze various mechanisms underlying COVID-19. Moreover, the 14 applications enable researchers to customize grouping and setting for different analyses and allow them to perform analyses using their own data. Furthermore, a Drug Discovery tool is designed to identify potential drugs and targets at whole transcriptomic scale. For proof of concept, we used COVID19db and identified multiple potential drugs and targets for COVID-19. In summary, COVID19db provides user-friendly web interfaces to freely analyze, download data, and submit new data for further integration, it can accelerate the identification of effective strategies against COVID-19. (website:http://hpcc.siat.ac.cn/covid19db/)
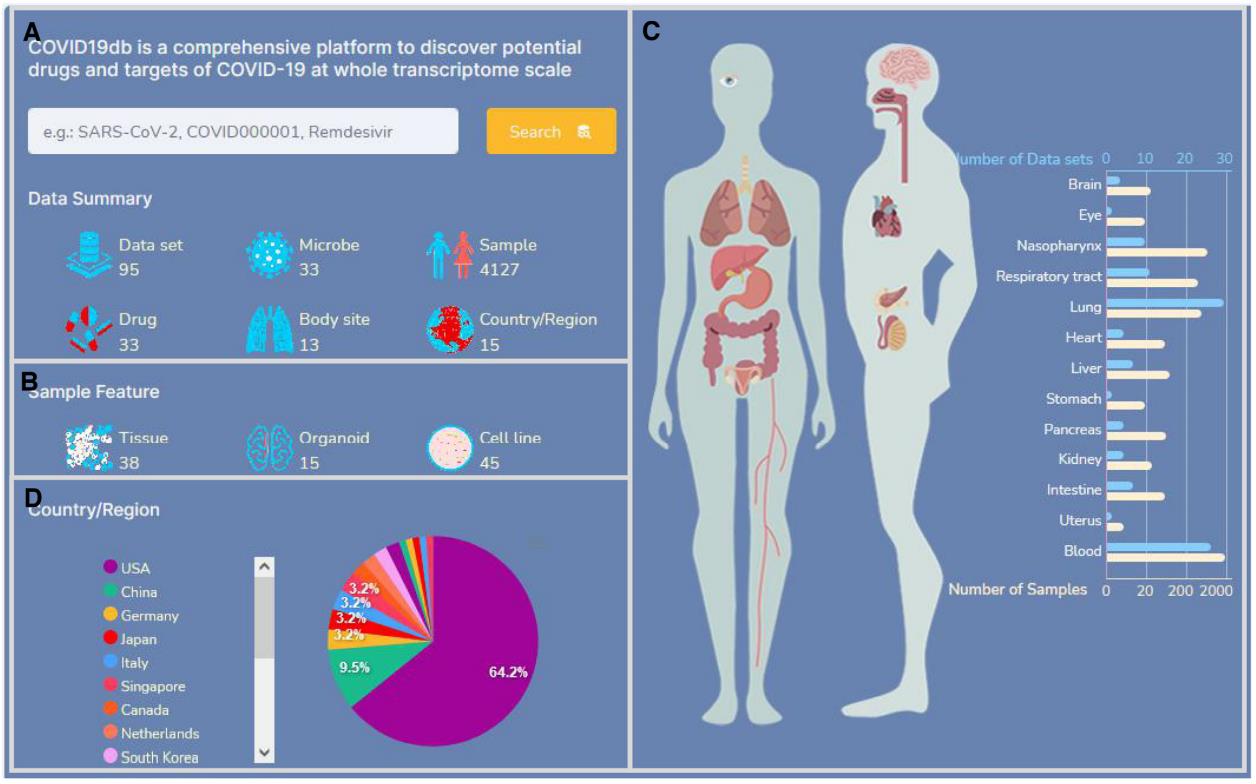 The data landscape and web interface of COVID19db
The data landscape and web interface of COVID19db