

IPS Cell Identification
(1) Machine Learning for iPS Progenitor Cell Identification. Identification of induced pluripotent stem (iPS) progenitor cells, the iPS forming cells in early stage of reprogramming, could provide valuable information for studying the origin and underlying mechanism of iPS cells. However, it is very difficult to identify experimentally since there are no biomarkers known for early progenitor cells, and only about 6 days after reprogramming initiation, iPS cells can be experimentally determined via fluorescent probes. What is more, the ratio of progenitor cells during early reprograming period is below 5%, which is too low to capture experimentally in the early stage. We propose a novel computational approach for the identification of iPS progenitor cells based on machine learning and microscopic image analysis. Our model allows several missing values/frames in the sample datasets, thus it is applicable to a wide range of scenarios.
 Flow chart of the machine learning based approach for iPS progenitor cell identification.
Flow chart of the machine learning based approach for iPS progenitor cell identification.
(2)A weakly supervised method for automatic cell detection and tracking. Automatic cell detection in microscopy image sequences is a significant yet challenging task in biomedicine. Compared with traditional methods based on feature extraction operators, convolutional neural network (CNN) based cell detection methods can achieve better performance. However, existing CNN-based methods requires manual annotations for model training, which is often time-consuming and labor-intensive. We propose a weakly-supervised cell detection method to train a convolutional neural network without manual annotations. The initial pseudo ground truth (label) was obtained from the fluorescence or bright field images through image processing techniques, and data augmentation was used to expand the sizes of the training sets. The label quality and detection performance was improved iteratively. For each iteration, a U-net based CNN architecture was trained for cell detection, coupled by a pseudo ground truth updating process using cell tracking technique. Evaluated on two independent datasets, the proposed method showed DET of 0.82- 0.92 for different iPS reprogramming time periods of our induced pluripotent stem (iPS) dataset using pseudo-labels generated from low-quality fluorescent images, and achieved DET of 0.84-0.98 for Fluo N2DH GOWT1 dataset from Cell Tracking Challenge using only 10%-90% of cell markers of reference annotations.
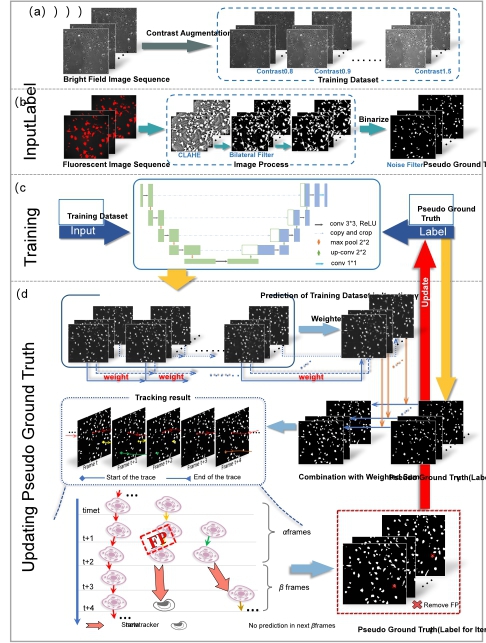 A weakly supervised method for automatic cell detection and tracking
A weakly supervised method for automatic cell detection and tracking